LSTM Models for Time Series (Stock Market Dataset)
LSTM (Long Short-Term Memory) models is a type of recurrent neural
network (RNN). Recurrent neural networks use the output of the previous layer as the input, and they can retain information about previous time steps in the sequence.
This works really well for time series because we are taking into account previous layers, and therefore, the time aspect of the data, its history.
Retrieving Data
We have a stock market dataset that contains values from 2010 to 2019. The training dataset contains the values
from 2010 to 2018.
And the testing dataset contains values from 2018 to 2019. The features are the following :
Date, Open, High, Low, Close, Adj Close, Volume.
We are only interested here in the Open values.
The training dataset and the testing dataset are in separate csv files. We will use pandas to read the csv
files.
(Link to get the two csv files)
import pandas as pd
data_train = pd.read_csv('training_data.csv')
data_test = pd.read_csv('test_data.csv')
data_train Date Open High Low Close Adj Close Volume
0 2010-07-28 37.667141 37.998573 37.178570 37.279999 32.658363 129996300
1 2010-07-29 37.244286 37.521427 36.585712 36.872856 32.301701 160951700
2 2010-07-30 36.555714 37.099998 36.414288 36.750000 32.194061 112052500
3 2010-08-02 37.205715 37.512856 37.088570 37.407143 32.769741 107013900
4 2010-08-03 37.287144 37.608570 37.060001 37.418571 32.779743 104413400
This is head of the training dataset.
Data Preparation
We are going to be retrieving the second column of the training dataset and the testing dataset.
data_train = data_train.iloc[:,1:2].values
data_test = data_test.iloc[:,1:2].valuesLet's visualize data_train.
import matplotlib.pyplot as plt
plt.plot(data_train)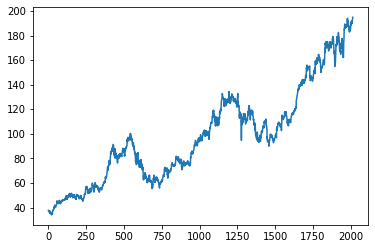
This is what data_train looks like. We can see that we have some trend and seasonality in the dataset.
For data preparation, we are going to start by scaling our data. This is very important for time series to capture data properly at different time scales. We are making sure that each observation is treated equally.
We will be using MinMaxScaler from scikit-learn library. If we don't specify the range of the scale, then by default the range will be (0,1).
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaler.fit(data_train)
data_train_scaled = scaler.transform(data_train)
data_test_scaled = scaler.transform(data_test)
data_train_scaled array([[0.022744 ],
[0.02011732],
[0.01584005],
...,
[0.98801121],
[0.9976395 ],
[1. ]])
For the final and most important step, we will be creating samples, reshaping our dataset and creating
x_train, y_train from data_train and x_test, y_test from data_test.
The idea behind this is to make the data into the following format :
# Let's say this is our data
data_train = [ [10], [7], [35], [20], [9], [2], [17], [85] , ... ]
t_step = 3
# Creating x_train, y_train from data_train with a "sliding windows", t_step=3
x_train = [ [[10], [7], [35]], [[7], [35], [20]], [[35], [20], [9]], [[20],[9],[2]],
[[9],[2],[17]] ... ]
y_train = [ [20], [9], [2], [17], [85] ... ]
What is above is just an example. Our data will not be looking like that because, remember, we have scaled it.
But this is the transformation that we are doing to be doing to our datasets.
For train_data
m_train, n_train = data_train.shape
m_test, n_test = data_test.shape
t_step = 60
y_train = list()
x_train = list()
for i in range(t_step,m_train):
# Create temporary samples
sample_x_train = data_train_scaled[i-t_step:i]
sample_y_train = data_train_scaled[i]
# Let’s add a padding
if sample_x_train.shape[0] < t_step:
var = np.zeros((t_step - sample_x_train.shape[0]),1)
sample_x_train = np.concatenate((sample_x_train, var), axis = 0)
sample_y_train = np.concatenate((sample_y_train, var), axis = 0)
# Adding to the lists x_train, y_train
x_train.append(sample_x_train)
y_train.append(sample_y_train)
x_train = np.array(x_train)
y_train = np.array(y_train)For test_data
t_step = 60
y_test = list()
x_test = list()
for i in range(t_step,m_test):
# Create temporary samples
sample_x_test = data_test_scaled[i-t_step:i]
sample_y_test = data_test_scaled[i]
# Let’s add a padding
if sample_x_test.shape[0] < t_step:
var = np.zeros((t_step - sample_x_test.shape[0]),1)
sample_x_test = np.concatenate((sample_x_test, var), axis = 0)
sample_y_test = np.concatenate((sample_y_test, var), axis = 0)
# Adding to the lists x_test, y_test
x_test.append(sample_x_test)
y_test.append(sample_y_test)
x_test = np.array(x_test)
y_test = np.array(y_test)Now that our data is preprocessed, let's create the LSTM model!
LSTM model
model = Sequential()
model.add(LSTM(units = 50, activation = "tanh", return_sequences = True, batch_size
= (None, t_step, 1))) #output 50
model.add(Dropout(0.1)) #dropout rate of 0.1
model.add(LSTM(units = 50, activation = "tanh", return_sequences = False))
model.add(Dropout(0.1))
model.add(Dense(units = 1)) #output layermodel.compile(loss='mse', optimizer='adam')
model.fit(x_train, y_train, epochs=100)
model.evaluate(x_test, y_test)Our model has been created! You can play around with these parameters to decrease the loss of the model and reduce errors between y_test and y_pred. To predict y_test, we can use :
y_pred = model.predict(y_test)And for the errors between y_test and y_pred :
import tensorflow as tf
tf.keras.metrics.mean_squared_error(y_test, y_pred)Results
We can plot the results so that we can notice the difference between the predictions and the real value
plt.plot(y_test)
plt.plot(y_pred)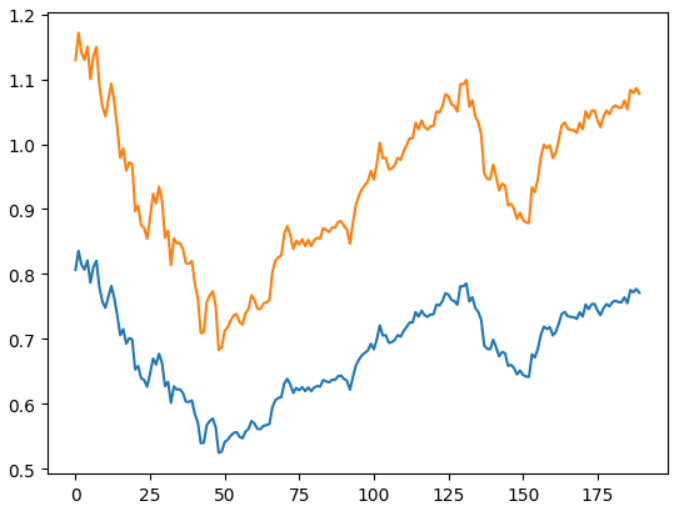
plt.plot(y_test,y_pred)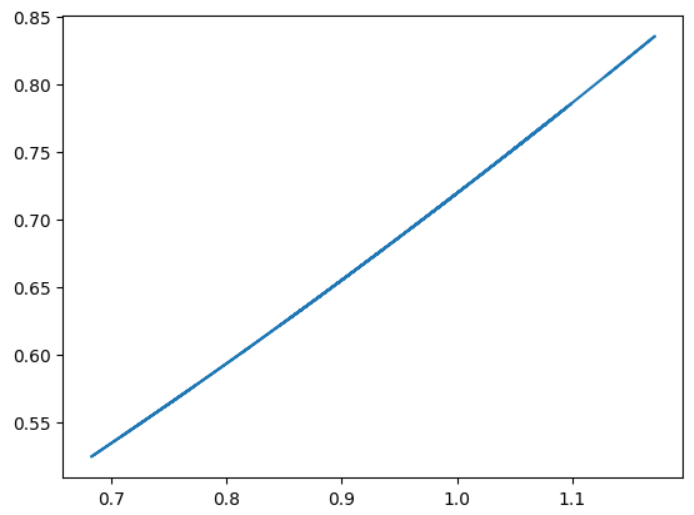
The results are really not that bad. The model is able to capture important
patterns and relationships in the data, like the trend and the seasonality. And it is very much close to the real values as we can see.
As for the second plot, the idea is to simply to plot it as x=y and to see the linearity if it exists and in this case
it kind of does!
We can also try to
There may still be room for improvement for this model, but we will stick with this for now.
If you have any questions or wants to know more about the project, feel free to email me or tweet at me!
Related : Introduction To Machine Learning
Join my newsletter for similar articles and early access